Leslie A. Lyons
Department of Population Health & Reproduction, School of Veterinary Medicine, University of California, Davis
Cat Diversity and Domestication
Despite the fact that nearly 70 million cats inhabit US households (U.S. Pet Ownership & Demographics Sourcebook, AVMA, 2002), little is known about the origins of the domestic cat. Equally scarce is information regarding the origins of the various cat breeds and their relationship to each other. The cat was more recently domesticated than most other companion animals and livestock species, but no molecular studies have been focused on deciphering domestic feline origins. The popularity of cats in Egyptian societies has lead to the assumption that cats were domesticated in Africa. Archeological evidence and early writings, however, place domestic cats in Asia and Eastern Europe prior to the rise of the cat in the Egyptian Empire. Recently, intact remains of a cat were identified at a Neolithic human burial site in Cyprus, which was dated at approximately 9,000 years ago, 5,000 years prior to the rise of cats in Egyptian culture{1}. Our objectives are to determine the domestication site of the cat, the relationship of wildcat populations to modern populations and breeds, and the population structure of modern cat breeds. We hypothesized that the cat was initially domesticated by civilizations in the Fertile Crescent, as a means of rodent control for crop stores and refuse sites.
Possible Progenitors: the Wildcats
The wildcat (Felis sylvestris Schreber, 1777) is commonly defined by four subspecies{2}, the domesticated subspecies (F. s. catus Linnaeus, 1758), and the African (F. s. libyca Foster, 1780), European (F. s. sylvestris) and Asian (F. s. ornata Gray, 1830) wildcats, however, many other sub-species have been noted and due to their widespread distribution, the taxonomy of wildcats is still under debate although domestication events have been proposed. The phylogeny of felids{3-5}. has been investigated, however, these studies did not focus on the most recent relationships of modern domestic cats to their progenitors. More recent morphological and genetic analyses have shed some light on the relationships of small European and African wildcats, and wildcats to domesticated cats, although the major focus of these studies have been on introgression and hybridization of wildcats with domestic cats and the resulting concerns with wild population conservation.
Cats in Archeology
Skull and jaw dimensions are currently the most distinguishing features between wild and domestic cat{6}, but many paleontological sites do not have sufficient remains for definitive recognition. Europe has several Pleistocene sites with small cat remains(1). These remains are considered to be wildcats because they are found among other non-domesticated carnivore remains. Teeth were found in Jericho, dated to ~9000 BC and ~4000 BC in the Indus Valley (1) but distinctions between wild or domestic cat could not be made. More recent archaeological material has been recovered from sites where suggestions of domestication are already apparent. Intact remains of a cat were identified at a Neolithic human burial site in Cyprus, which was dated at approximately 9,000 years ago, 5,000 years prior to the rise of cats in Egyptian culture. The discovery of skeletal remains consistent with the size of F. sylvestris, dating from ~7,000 BC on Cyprus, implies that cats were traveling on ships with humans. Furthermore, the ritualistic positioning of the cat next to a human burial suggests possible cultural or religious importance of cats to the people inhabiting Cyprus at that time.
Egyptian feline remains are abundant and well documented{7}. The earliest cat representations and mummies date to the 12th Dynasty (1991 - 1778 BC). Cats became a very popular entombment companion in the Ptolemaic Dynasty, ~320 BC, and were specifically bred for mummification.
Artwork and writings show strong evidence that the cat began to have a role in many societies, perhaps independently, throughout the Old World(1,20). Sanskrit writings from India describe small cats as early as ~3000 BC. Chinese cultural works present cats at ~2000 BC with clear domestication circa 400 AD. Domestic cats are considered to have entered Japan at ~1000 AD from China. In Greece, the appearance of cats in artwork is evident at 1500 BC. Thus, Egyptians may have domesticated cats and/or domesticated cats may have been brought to Egypt via trade routes.
Contemporary Domestication: Development of Modern Breeds
Historical information of modern breed development identifies several once geographically-isolated populations that represent the earliest divergences of the domestic cat species, F. catus, and/or possible introgressive hybrids between F. catus and F. sylvestris. Darwin noted in 1868 that cats from South America and India were different from those found in England(21). Abyssinians were disputed to have been Ethiopian- or British-derived; and Persians were putatively described as having Pallas cat progenitors, F. manul(22). The first cat show was held at the Crystal Palace in England in 1871. At this time, British, Persian, Manx, Siamese, Angora and Russian Blue breeds were highly recognizable and distinctive. These early breeds were recognized prior to establishment of the National Cat Club in 1887, which initiated strong and formal breeding practices, thus they represent geographical distinctions. Along with the Maine Coon, an American foundation breed, these breeds were the first cats to be presented at shows in the United States. As the cat fancy has increased in popularity over the past century, the number of defined breeds has also increased(22) Therefore, some breeds that have been recognized over the past 100 years may truly represent geographical isolates while others do not. Some breeds are clearly new mutations that have occurred in recognized populations. Many recently designated breeds have been produced by creative husbandry practices. There are over forty recognized cat breeds, approximately 50% of these can be identified as potential geographical isolates, useful for estimating ancient migration patterns and gene flow. These isolated breeds may reflect several different domestication events, or they may trace to only one event from a single small wildcat species.
Mitochondrial DNA
Due to its non-recombining nature and fast mutation rate mitochondrial DNA is the perfect molecule for studying genealogies of recently diverged populations, such as the domestic cat breeds. We have successfully amplified a 474 bp fragment of the 5' portion of the mtDNA control region in domestic breeds and random bred populations. To date, 149 individuals from 17 breeds and 90 random bred cats have been sequenced. Thirty different haplotypes have been identified, twelve haplotypes were unique to a breed and not found in the random bred population (Table 1). However, the random bred population represents cats from only southern California. Nine haplotypes were unique to the random bred cats, although this number is likely to be reduced as more pure bred cats are sampled. Three breeds had more than one unique haplotype. Overall, the preliminary data shows a large amount of diversity in both random bred cats and pure bred cats. The Abyssinian breed, however, seems to have low diversity despite its reputed ancient origins and relatively large population.
Table 1. Mitochondrial DNA haplotypes of cats breeds and random breds.
|
Breed |
N |
Haplotypes |
Breed |
N |
Haplotypes |
|
Abyssinian |
13 |
26, 29 |
Norwegian Forest Cat |
16 |
6, 17, 26, 29 |
|
American Shorthair |
10 |
9, 25, 26 |
Persian |
10 |
6, 7, 17, 26 |
|
Birman |
10 |
29 |
Russian Blue |
5 |
6, 26, 29 |
|
British Shorthair |
6 |
26 |
Siamese |
14 |
5, 6, 15, 16, 26, 29 |
|
Chartreux |
4 |
6, 29 |
Scottish Fold |
2 |
1 |
|
Egyptian Mau |
2 |
29 |
Selkirk Rex |
1 |
10 |
|
Japanese Bobtail |
11 |
8, 20, 26, 29 |
Turkish Angora |
2 |
11, 20 |
|
Korat |
9 |
2, 29 |
Turkish Van |
4 |
13 |
|
Maine Coon |
25 |
14, 22, 26, 27, 29 |
|
|
|
|
Random Bred |
90 |
3, 4, 5, 6, 10, 12, 18, 19, 20, 21, 24, 26, 28, 29, 30 |
Haplotypes in bold are unique to that breed/population.
A neighbor-joining tree (Figure 1) was constructed using a subset of the samples. As expected, individuals from some of the older foundation breeds, such as the Persian and Russian Blue, are present in more than one cluster thus exhibiting more genetic diversity. Another observation is that all of the Turkish Angoras examined to date have the same, apparently unique, haplotype. This observation agrees with the historical account of the breed as being a small, geographically isolated population. More interesting insights are expected to emerge as more individuals, especially the different subspecies of wildcats, are added to the analysis.
Microsatellites
Microsatellite markers have long been used in population based studies and have recently proven to be useful in distinguishing between breeds{8}. These markers mutate at very high rates and therefore are useful when studying recent diversification events. Surveying 20 breeds and several random bred populations, we have found there to be high levels of genetic variation. The PIC values range from 0.5 - 0.86, and private alleles are present in many populations (data not shown).
Phylogenetic analysis on the microsatellite data has shown some interesting trends as pertaining to cats of African origin. A Neighbor-Joining tree constructed using Nei's genetic distance groups the feral cat population from the islands of Faza and Lamu (off the coast of Kenya) with both the African and European Wildcats, supporting the theory of the ancient origins of these island cats. A subsequent analysis using a Bayesian clustering algorithm as implemented in the software Structure was performed{9}. Here the Lamu cats were analyzed separately from the cats on the neighboring island of Faza. Seven of the ten Lamu cats analyzed formed a strong cluster along with a single Sokoke cat. Historically the Sokoke breed is said to have originated from feral Kenyan cats and our results support the theory that at least some of the cats in this breed show strong association with feral African cats.
| Figure 2. | 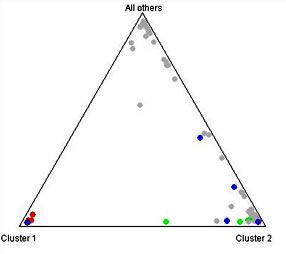
Triangle plot. Lamu cats are in red, Sokoke in blue, and Faza in green. All other cats are in grey. |
|
| |
Conclusion
Our preliminary results clearly show the usefulness of both mitochondrial DNA and microsatellite markers as tools for elucidating the genetic structure and genealogy of such recently diverged groups as domestic cat breeds. Our future efforts are focused on expanding our samples and genetic markers. One genetic marker system of interest is the Y chromosome. Our exploration of the genetic information contained on the cat Y chromosome has been slowed by the poor understanding of the Y chromosome in most companion animals.
References
1. Vigne, J.D., et al., Early taming of the cat in Cyprus. Science, 2004. 304(5668): p. 259.
2. Nowell, K. and P. Jackson, Wild Cats. 1996: IUCN/SSC Cat Specialist Group.
3. Johnson, W.E. and S.J. O'Brien, Phylogenetic reconstruction of the Felidae using 16S rRNA and NADH-5 mitochondrial genes. J Mol Evol, 1997. 44 Suppl 1: p. S98-116.
4. Johnson, W.E., et al., Resolution of recent radiations within three evolutionary lineages of Felidae using mitochondrial restriction fragment length polymorphism variation. J. Mamm. Evol., 1996. 3: p. 97-120.
5. Masuda, R., et al., Molecular phylogeny of mitochondrial cytochrome b and 12S rRNA sequences in the Felidae: ocelot and domestic cat lineages. Mol Phylogenet Evol, 1996. 6(3): p. 351-65.
6. Yamaguchi, N., et al., Craniological differentiation between European wildcats (Felis sylvestris sylvestris), African wildcats (F. s. lybica) and Asian wildcats (F. s. ornata): implications for their evolution and conservation. Biological Journal of the Linnean Society, 2004. 83: p. 47-63.
7. Malek, J., The cat in Ancient Egypt. 1993, Philadelphia: University of Pennsylvania.
8. Parker, H.G., et al., Genetic structure of the purebred domestic dog. Science, 2004. 304(5674): p. 1160-4.
9. Pritchard, J.K., M. Stephens, and P. Donnelly, Inference of population structure using multilocus genotype data. Genetics, 2000. 155(2): p. 945-59.